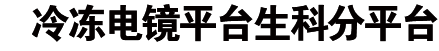Structures of SARS-CoV-2 B.1.351 neutralizing antibodies provide insights into cocktail design against concerning variants
The spread of the SARS-CoV-2 variants, especially the global variants of concern (VOCs), could seriously dampen our efforts to tackle the COVID-19 pandemic. The SARS-CoV-2 spike protein recognizes the host angiotensin-converting enzyme 2 (ACE2) via its receptor-binding domain (RBD) to mediate viral entry into the cells. Several notorious mutations have been identified in the spike RBD of the VOCs. For example, B.1.1.7 (Alpha), B.1.351 (Beta), and P.1 (Gamma) all contain the N501Y mutation, which increases the binding affinity for human ACE2 and confers higher infectivity in mice.1,2 In addition, B.1.351 and P.1 carry the K417N/E484K and K417T/E484K substitutions, respectively, which drastically alter RBD surface electrostatics and lead to immune escapes.3,4 E484K has been detected in some strains of B.1.1.7 as well. Recently, the World Health Organization classified B.1.617.2 (Delta) as the fourth global VOC, which contains the L452R and T478K double mutations in the RBD. The L452R substitution has been shown to resist some neutralizing antibodies,5,6 and is present in other SARS-CoV-2 variants besides B.1.617.2, such as B.1.617.1 (Kappa) and B.1.427/B.1.429 (Epsilon). A L452Q mutation is found in C.37 (Lambda). The functional significance of T478K remains to be understood.
https://www.nature.com/articles/s41422-021-00555-0