Structures of a P4-ATPase lipid flippase in lipid bilayers
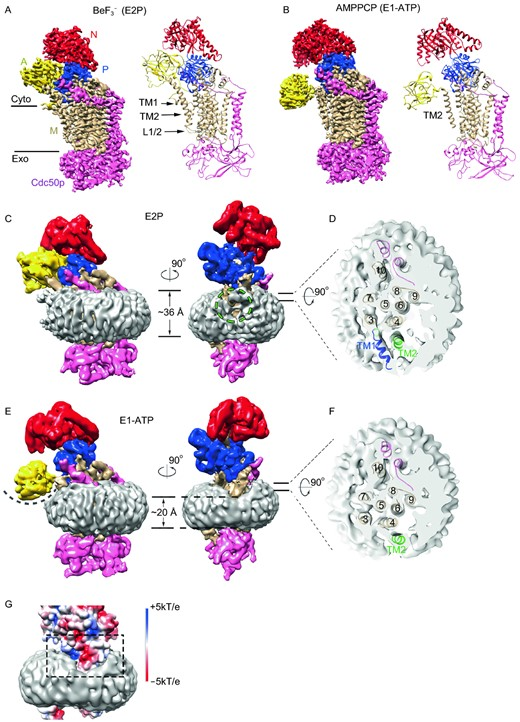
Phospholipid molecules are unevenly distributed in membrane bilayers of eukaryotic cells. Phosphatidylethanolamine (PE) and phosphatidylserine (PS) are concentrated in the cytoplasmic leaflet, whereas phosphatidylcholine (PC) is enriched in the exoplasmic leaflet (lumenal or extracellular leaflet) (van Meer et al. 2008). Type 4 P-type ATPases (P4-ATPases) are the phospholipid flippases which move specific lipids from the exoplasmic leaflet to the cytoplasmic leaflet. P4-ATPases belong to the P-type ATPase family. P4-ATPases are conserved in eukaryotes. Saccharomyces cerevisiae (S. cerevisiae) has five P4-ATPases characterized, namely Drs2p, Dnf1p, Dnf2p, Dnf3p, and Neo1p, whereas 14 P4-ATPases are identified in human. Each P4-ATPase has its preferred lipid substrates. For example, yeast Drs2p translocates PS and PE, whereas Dnf1p and Dnf2p prefer PC, PE, and glucosylceramide (Roland et al. 2019). Most P4-ATPases form heterodimers with a β-subunit from the CDC50 family. The β-subunit is required for proper folding, sub-cellular targeting, and lipid flipping of P4-ATPases (Radji et al. 2001; Bryde et al. 2010). Lipid flipping by P4-ATPases is coupled with ATP hydrolysis and enzyme phosphorylation/dephosphorylation. P4-ATPases undergo the E1–E2 state transition during lipid flipping cycles. Lipid substrate binding is coupled with dephosphorylation in the low energy E2P state whereas the role of the E1 state in lipid flipping is less clear. The lipid flipping pathway in P4-ATPases is also under debate. A “two-gate” model suggests that specific phospholipids are recognized at the entry and exit gates by the residues clustering at the exoplasmic and cytoplasmic ends of transmembrane segments (TMs) 1–4. The polar heads of the lipid substrates could slide through a groove formed by TMs 1, 3, and 4 during flipping (Baldridge and Graham 2013). A “hydrophobic gate” model proposed a cross-membrane groove bordered by TMs 1, 2, 4, and 6. A highly conserved isoleucine residue (I364 in bovine ATP8A2) and a few hydrophobic residues nearby act as a hydrophobic gate to control lipid movements (Vestergaard et al. 2014).
https://academic.oup.com/proteincell/article/11/6/458/6746812